Observations of marine biology
- This page is part of the topic Observations, data accuracy and tools
From historical surveys to high-tech instrumentation
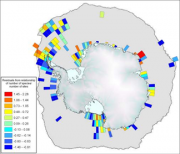
Historical expeditions in Antarctic offshore waters dating back to the second half of the 19th century can provide data relevant to studies of the effects of climate on the ecosystem, and can provide a baseline for long-term observations, as has been demonstrated in studies of krill and salp populations (Atkinson et al., 2004[2]). Early taxonomic surveys and corresponding species descriptions provide much of the basis for advanced studies on ecosystem functioning, especially where historical taxonomic and biogeographic data are included. Such data are generally accessible through databases managed by by SCAR-MarBIN, SOMBASE (Figure 2.29), FISHBASE, OBIS or, for a more multidisciplinary approach, by PANGAEA.
Modern taxonomic and biogeographic surveys include programmes such as CAML (Census of Antarctic Marine Life) and ICEFISH (International Collaborative Expedition to collect and study Fish Indigenous to Sub-Antarctic Habitats), both linked to the SCAR/IPY biology program EBA (Evolution and Biodiversity in the Antarctic – The Response of Life to Change). Long-term observations are recognised by an increasing number of scientists as an essential tool not only to achieve sustainable yields of natural resources such as krill and fish and to study the impact of over-exploitation e.g. by CCAMLR surveys (Convention on the Conservation of Antarctic Marine Living Resources), but also to monitor the impact of climate change at any level of ecosystem organisation.
Satellite sensors, combined with ‘ground truth’ data from in situ surveys, are contributing to a better understanding of pelagic systems by providing large scale and long-term data on biological bulk parameters such as chlorophyll, and on ecologically relevant physical parameters, such as ice cover, sea surface temperature, and iceberg production. ADCP’s and plankton-video-recorders provide further information on the pelagic habitat to supplement results from traditional net hauls and CTD stations. Moored sediment traps and current meters provide data on the pelagic environment and on biological coupling between the surface and the seabed. High resolution side scanning sonars, benthic landers, cameras and ROVs provide further information. These methodologies provide the basis for classifying the benthic system and for carrying out controlled sampling, for instance of sediments and organisms at a cold seep in the former Larsen B ice shelf area, for observing small scale patterns of recolonisation after ice berg scouring, and for carrying out fine-scale experiments on the effects of increased supplies of planktonic phytodetritus as a source of food for the sea-floor.
Continuous recording of underwater acoustics, e.g. from the year-round operating PALAOA observatory beneath the Ekström Ice Shelf, provides information on the activities of whales and seals that can be used to study aspects of the ecology of the top predators and of nature conservation (Boebel et al., 2008[3]).
In the future, the routine use of AUVs and gliders will provide a picture at even higher spatial resolution of ocean behaviour and ecology, in addition to permitting collection of data from formerly inaccessible areas under the sea ice or beneath ice shelves. Instrumented AUVs could also contribute to comprehensive mapping of sea-floor habitats as the basis for characterising seabed topography and sediments for classifying benthic communities surveying krill and other planktonic concentrations and, finally, establishing ecosystem function. Fine scale experiments and observations can be continued through the use of instrumented ROVs and the invention of independently operating crawlers triggered by events. Automatically operating underwater observatories can be used to measure in situ a variety of ecologically relevant parameters for detecting biological response to climate change and for large scale ecological mapping.
Intensive inshore biological research supported by land stations began in the 1960s. Even at that early stage, sampling was accompanied by simple but effective in situ experiments carried out by scuba divers. Later, complicated laboratory experiments were made possible by new opportunities to maintain living animals in aquaria with running sea-water, not only on bases on the Antarctic continent, but also on research vessels carrying fish and invertebrates to home institutes. Such physiological studies contribute especially to a better understanding of the presence and future of marine biota if they are combined with field obervations and field surveys. Modern station-based and scuba-diving-based ecosystem field research focuses on determining the ecological effects of retreating and melting glaciers and ice scouring at the organism, species and community levels, for instance at Rothera, McMurdo, and Jubany Stations.
Molecular biology, genetics, genomics
The role of molecular analyses is exponentially growing. A new powerful aid to evolutionary biology and ecology has been recently provided by genomic technologies, whose fast development has brought biological sciences to the threshold of a revolution. The availability of sequences greatly enhances our understanding of the structure, function, content, variation and evolution of individual genes. Most importantly, it allows sequencing of whole genomes. Sequencing genomes of model organisms is a great challenge for biological sciences. In the last decade, scientists have developed methods to align and compare sequenced genomes, but the analysis of a given sequence provides much information on the genome structure, yet only to a lesser extent on the function. Comparative genomics is a useful tool for functional and evolutionary annotation of genomes. In principle, comparison of genomic sequences may allow for identification of evolutionary selection (negative or positive) the functional sequences have been subjected to over time. Positively selected genome regions are the most important ones for evolution, because most changes are adaptive and often induce biological differences in organisms.
Genome mining is clearly high on the agenda and it also provides unparalleled tools for studying natural selection in action, and investigating the link between organisms and environment (environmental genomics). DNA sequence data on Antarctic bacteria are increasingly available. Genomics will provide invaluable information on the molecular basis of adaptation to the extreme marine environment at the poles, and on the response to climate changes. DNA microarrays will lead to high throughput analysis of gene expression (functional genomics) in marine polar organisms. Structural genomics, i.e. sequencing genomic libraries (bacterial artificial chromosome (BAC) libraries) and whole-genome sequencing, would complement the information from transcriptomics, to understand how the extreme polar environment has shaped genome evolution. Genome sequencing will also provide the means for a population genomic (rather than genetic) approach to the study of gene flow, population structure and dynamics of marine polar organisms. Large sequencing projects generally exceed the capacity of single research groups and even national programmes, and therefore cooperative international actions are highly desirable. An additional goal will be to make marine biologists, ecologists, physiologists, and other polar scientists familiar with genomics. The overall objective is to review and coordinate current knowledge of gene/protein evolution, adaptations and systematics (proteomics) in polar marine organisms.
Genomics is high-profile science, impacting on all areas of biology, so it is not surprising that it is playing an increasing role in polar studies (Clark et al., 2004[4]). Future research in genomics will not only depend on the new technologies, but also on the integration with ecology, physiology and biochemistry.
Nowadays molecular biology is essential for studying evolution, and important insights come from nucleic acid and protein sequences, as well as from large-scale chromosome change (e.g. Stam et al., 1997[5], 1998[6]; di Prisco, 1998[7], 2000[8]; Bargelloni et al., 2000a[9]; Pisano et al., 1998[10], 2003[11]; Held, 2000[12]; Pawlowski et al., 2002[13]; Papetti et al., 2007[14]; Wilson et al., 2007[15]; Mahon et al., 2008[16]; Smith et al., 2008[17]; Strugnell et al., 2008[18]). Relevant examples of molecular phylogeny, e.g. hemoglobin, antifreeze glycoprotein, myoglobin (Cheng, 1998[19]; Bargelloni et al., 2000b[20]; Near, 2004[21]; Verde et al., 2006a[22],b[23]; Giordano et al., 2007[24]), have expanded our knowledge on adaptive evolution, mostly related to the fish realm.
Cold adaptation is an important part of a refined physiological equilibrium, which remains unmodified in the absence of disturbances due to climate change. When such disturbances do occur, evaluating the response of cold-adapted organisms will yield indications which can be extrapolated to organisms of lower latitudes.
Knowledge on the capacity of the polar marine fauna to respond to on-going climate changes is beginning to emerge, e.g. information is available on fish cold adaptation. Over the last twenty years, important advances have been made in understanding the molecular mechanisms involved in evolutionary adaptation to temperature change. Further progress is likely to occur as scientists exploit new molecular approaches to environmental changes in the polar regions. Increasing knowledge of protein structure and function is progressively helping to solve the puzzle of cold adaptation. Although in some proteins a coherent picture of the molecular changes involved in evolutionary adaptation to temperature is emerging, much remains unknown as yet. Relatively few cold-adapted enzymes have been examined, and many important cellular processes have hardly been studied at all. Nowadays, the possibility of sequencing whole genomes may provide the amount of data needed to allow rejection or acceptance of some of the classical hypotheses currently invoked to explain protein thermal adaptation.
The use of molecular techniques in diversity, phylogenetic and population research has opened new ways to analyse patterns in biogeography and evolution. DNA markers, for example mitochondrial genes (e.g. encoding cytochrome oxidase subunit I) 12S and 16S rDNA, or nuclear genes 18S and 28S rDNA, rhodopsin or pax-6, can be used to identify species and to analyse macroevolution (phylogenetics – studying genome mutations over time – and phylogeography) and microevolution (population genetics – studying relative frequency of genome variation and how they change over time). In the past decade several molecular studies on Antarctic marine and terrestrial fauna have identified the existence of cryptic (hidden) species (e.g. Held and Wägele, 2005[25]; Linse et al., 2007[26]; Raupach and Wägele, 2006[27]; Brökeland and Raupach, 2008[28]). Cryptic species are morphologically very similar but vary in their nucleic acid sequences. Since its introduction, DNA barcoding has been used to identify species (e.g. Webb et al., 2006[29]; Rock et al., 2008[30]) and to reveal new species (e.g. Lörz et al., 2007[31]; Smith et al., 2008[17]), mostly using the COI gene.
A robust knowledge of species distributions is important as it gives insights into Antarctica’s faunal and floral past across time scales from recent ecological scale disturbance (e.g ice scour in marine habitats) through glacial cycles to the fragmentation of Gondwanaland (Convey et al., 2008[32], in review). Recent phylogeographic studies have provided an insight as to the origins and maintenance of current Antarctic diversity by examining how biota have responded to past and current climate and other environmental fluctuations (e.g. Raupach et al., 2004[33]; Stevens and Hogg, 2003[34]; Stevens et al., 2006[35]; Allegrucci et al., 2006[36]; Van Vuuren et al., 2007[37]; Wilson et al., 2007[15]). Calibrating phylogenies with fossil records reduces the error in molecular clock estimates and allows a tighter comparison between inferences from molecular data and the historical climate and geological records (e.g. Page and Linse, 2002[38]; Strugnell et al., 2008[18]).
References
- ↑ Griffiths, H.J., Linse, K. and Crame, J.A. 2003. SOMBASE – Southern Ocean Mollusc Database: a tool for biogeographic analysis in diversity and ecology, Organisms Diversity and Evolution, 3, 207-213.
- ↑ Atkinson, A., Pakhomov, E., Rothery, P., Siegel, V. 2004. Long-term decline in krill stocks and increase in salps within the Southern Ocean, Nature, 432 (7013), 100-103.
- ↑ Boebel, O., Klinck, H., Kindermann, L. and EL Nagar, S. 2008. PALAOA: Broadband recordings of the Antarctic cosatal soundscape, Bioacoustics, 17, 19-21.
- ↑ Clark, P.U., McCabe, A.M., Mix, A.C. and Weaver, A.J. 2004. Rapid rise of sea level 19,000 years ago and its global implications, Science, 304, 1141-1144.
- ↑ Stam, W.T., Beintema, J.J., D'avino, R., Tamburrini, M. and Di Prisco, G. 1997. Molecular evolution of Antarctic fishes (Notothenioidei), J. Mol. Evol., 45, 437-445.
- ↑ Stam, W.T., Beintema, J.J., D’Avino, R., Tamburrini, M., Cocca, E. and Di Prisco, G. 1998. Evolutionary studies on teleost hemoglobin sequences. In: di Prisco G, Pisano E and Clarke A (Eds) Fishes of Antarctica. A Biological Overview, 355-359. Springer, Milano.
- ↑ Di Prisco, G. 1998. Molecular adaptations in Antarctic fish hemoglobins. In: di Prisco G, Pisano E and Clarke A (Eds) Fishes of Antarctica. A Biological Overview, Springer, Milano, 339-353.
- ↑ Di Prisco, G. 2000. Life style and biochemical adaptation in Antarctic fishes, J. Mar. Syst., 27, 253-265.
- ↑ Bargelloni, L., Marcato, S., Zane, L. and Patarnello, T. 2000a. Mitochondrial phylogeny of notothenioids: a molecular approach to Antarctic fish evolution and biogeography, Syst. Biol., 49, 114-129.
- ↑ Pisano, E., Ozouf-Costaz, C. and Prirodina, V. 1998. Chromosome diversification in Antarctic fish (Notothenioidei). In: di Prisco G, Pisano E and Clarke A (Eds) Fishes of Antarctica. A Biological Overview, 275-285, Springer, Milano.
- ↑ Pisano, E., Cocca, E., Mazzei, F., Ghigliotti, L., Di Prisco, G., Detrich, H.W. III and Ozouf-Costaz, C. 2003 Mapping of alpha- and beta-globin on Antarctic fish chromosomes by fluorescence in situ hybridisation, Chromosome Res., 11, 633-640.
- ↑ Held, C. 2000. Phylogeny and biogeography of serolid isopods (Crustacea, Isdopoda, Serolidae) and the use of ribosomal expension segments in molecular systematics, Molecular Phylogenetics and Evolution, 15(2), 165-178.
- ↑ Pawlowski, J., Fahrni, J.F., Brykczynska, U., Habura, A. and Bowser S.S. 2002. Molecular data reveal high taxonomic diversity of allogromiid Foraminifera in Explorers Cove (McMurdo Sound, Antarctica), Polar Biol., 25(2), 96-105.
- ↑ Papetti, C., Liò, P., Rüber, L., Patarnello, T. and Zardoya, R. 2007. Antarctic fish mitochondrial genomes lack ND6 gene, J. Mol. Evol., 65, 519-528.
- ↑ 15.0 15.1 Wilson, N.G., Hunter, R.L., Lockhart, S.J. and Halanych, K.M. 2007. Multiple lineages and an absence of panmixia in the ‘circumpolar’ crinoid Promachocrinus kerguelensis in the Atlantic sector of Antarctica, Mar. Biol., 152, 895-904.
- ↑ Mahon A.R., Arango C.P. and Halanych K.M. 2008. Genetic diversity of Nymphon (Arthropoda: Pycnodonida: Nymphonidae) along the Antarctic Peninsula with a focus on Nymphon austral Hodgson 1902, Mar. Biol., 155, 315-323.
- ↑ 17.0 17.1 Smith, P.J., Steinke, D., McVeagh, S.M., Stewart, A.L., Struthers, C.D. and Roberts, C.D. 2008. Molecular analysis of Southern Ocean skates (Bathyraja) reaveals a new species of Antarctic skate, Journal of Fish Biology, 73, 1170-1182.
- ↑ 18.0 18.1 Strugnell, J.M., Rogers, A.D., Prodöhl, P.A., Collins, M.A. and Allcock, A.L. 2008. The thermohaline expressway: the Southern ocean as a centre of origin for deep-sea octopuses, Cladistics, 24, 853-860.
- ↑ Cheng, C-H.C. 1998. Origin and mechanism of evolution of antifreeze glycoproteins in polar fishes. In: di Prisco G, Pisano E, Clarke A (Eds) Fishes of Antarctica. A Biological Overview, Springer, Milano, 311-328.
- ↑ Bargelloni, L., Zane, L., Derome, N., Lecointre, G. and Patarnello, T. 2000b. Molecular zoogeography of Antarctic euphausiids and notothenioids: from species phylogenies to intraspecific patterns of genetic variations, Antarctic Sci., 12, 259-268.
- ↑ Near, T. 2004. Estimating divergence times of notothenioid fishes using a fossil-calibrated molecular clock, Antarctic Sci., 16, 37-44.
- ↑ Verde, C., Balestrieri, M., De Pascale, D., Pagnozzi, D., Lecointre, G. and Di Prisco, G. 2006a. The oxygen-transport system in three species of the boreal fish family Gadidae. Molecular phylogeny of haemoglobin, J. Biol. Chem., 281, 22073-22084.
- ↑ Verde, C., Giordano, D. and Di Prisco, G. 2006b. Molecular evolution of haemoglobins of polar fishes, Rev. Envir. Sci. Biotech., 5, 297-308.
- ↑ Giordano, D., Parrilli, E., Dettaï, A., Russo, R., Barbiero, G., Marino, G., Lecointre, G., Di Prisco, G., Tutino, L. and Verde, C. 2007. The truncated hemoglobins in the Antarctic psychrophilic bacterium Pseudoalteromonas haloplanktis TAC125, Gene, 398, 143-155.
- ↑ Held, C. and Wägele, J.W. 2005. Cryptic speciation in the giant Antarctic isopod Glyptonotus antarcticus (Isopoda: Valvifera: Chaertiliidae), Scientia Marina, 69(2), 175-181.
- ↑ Linse, K., Cope, T., Lörz, A.-N. and Sands C.J. 2007. Is the Scotia Sea a center of Antarctic marine diversification? Some evidence of cryptic speciation in the circum-Antarctic bivalve Lissarca notorcadensis (Arcoidea: Philobryidae), Polar Biol., 30, 1059-1068.
- ↑ Raupach, M.J. and Wägele, J.W. 2006. Distinguishing cryptic species in Antarctic Asellota (Crustacea: Isopoda) - a preliminary study of mitochondrial DNA in Acanthaspidia drygalskii, Antarct. Sci., 18, 191-198.
- ↑ Brökeland W. and Raupach M. 2008. A species complex within the isopod genus Haploniscus (Crustacea: Malacostraca: Peracarida) from the Southern Ocean deep sea: a morphological and molecular approach, Zoological Journal of the Linnean Society, 152, 655-702.
- ↑ Webb, K.E., Barnes, D.K.A., Clark, M.S. and Bowden, D.A. 2006. DNA barcoding: a molecular tool to identify Antarctic marine larvae, Deep-Sea Res. II,, 53 (8-10), 1053-1060.
- ↑ Rock, J., Costa, F.O., Walker, D.I., North, A.W., Hutchinson, W.F. and Carvalho, G.R. 2008. DNA barcodes of fish of the Antarctic Scotia Sea indicate priority groups for taxonomic and systematics focus, Antarct. Sci., 20, 253-262.
- ↑ Lörz A.-N., Maas E.W., Linse K. and Fenwick G.D. 2007. Epimeria schiaparelli sp nov., an epimeriid amphipod from the Ross Sea with a molecular characterization, Zootaxa, 1420, 23-37.
- ↑ Convey, P., Gibson, J.A.E., Hillenbrand, C-D., Hodgson, D.A., Pugh, P.J.A., Smellie, J.L. and Stevens, M.I. 2008. Antarctic terrestrial life - challenging the history of the frozen continent? Biological Reviews, 83, 103-117.
- ↑ Raupach, M.J., Held, C. and Wägele, J.W. 2004. Multiple colonization of the deep sea by the Asellota (Crustacea: Peracarida: Isopoda), Deep-Sea Research II, 51, 1787-1795.
- ↑ Stevens, M.I. and Hogg, I.D. 2003. Long-term isolation and recent range expansion revealed for the endemic springtail Gomphiocephalus hodgsoni from southern Victoria Land, Antarctica, Molecular Ecology, 12, 2357-2369.
- ↑ Stevens, M.I., Greenslade, P., Hogg, I.D. and Sunnucks, P. 2006. Southern hemisphere springtails: could any have survived glaciation of Antarctica?, Mol. Biol. Evol., 23, 874-882.
- ↑ Allegrucci, G., Carchini, G., Todiscso, V., Convey, P. and Sbordoni, V. 2006. A molecular phylogeny of Antarctic Chironomidae and its implications for biogeographical history. Polar Biology, 29, 320-326.
- ↑ Van Vuuren, B.J., Mortimer, E., Stevens, M.I., Marshall, D.J., Convey, P., Daniels, S.R. and Chown, S.L. 2007. Molecular data can help to unveil biogeographic complexities since the Miocene: lessons from ameronothroid mites and isotomid springtails – Online Proceedings of the 10th ISAES (http://pubs.usgs.gov/of/2007/1047/).
- ↑ Page, T.J. and Linse, K. 2002. More evidence of speciation and dispersal across the Antarctic Polar Front through molecular systematics of Southern Ocean Limatula (Bivalvia: Limidae), Polar Biol., 25, 818-826.